About the Laboratory
Newcastle Genetics Laboratory is the regional hub for rare disease genomic testing in the North East and North Cumbria, as part of the North East and Yorkshire Genomic Laboratory Hub (NEY GLH). The NEY GLH is laboratory network that provides comprehensive genomic services to a population of around 8–10 million across North East England and Yorkshire. Working in collaboration with partner laboratories in Leeds and Sheffield, our hub ensures equitable access to high-quality genetic testing for patients with rare inherited conditions. All testing is delivered in accordance with the NHS England National Genomic Test Directory, which defines the available tests, technologies, and patient eligibility criteria nationally.
Our laboratory is accredited for medical testing (ISO:15189), demonstrating our commitment to quality and patient safety. We work closely with the NHS Genomic Medicine Service (GMS) to integrate the latest genomic technologies into routine patient care. The Newcastle Genetics Laboratory provides rare disease and acquired cancer genomics services. We collaborate with clinical genetics teams and specialty clinics (such as the National Renal Complement Therapeutics Centre for atypical Haemolytic Uraemic Syndrome (aHUS) and the John Walton Muscular Dystrophy Centre for Limb Girdle Muscular Dystrophy (LGMD)) to deliver a seamless service from test ordering to results interpretation. Our service provides timely and accurate genetic diagnoses that inform patient management and family planning, improving outcomes for individuals with rare genetic disorders.
Our Rare Disease Genomic Services
We offer a comprehensive range of genomic tests for rare inherited diseases, spanning traditional chromosome analysis through to cutting-edge genome sequencing. All tests are aligned with the NHS National Genomic Test Directory, ensuring that the appropriate test is available for each clinical indication.
While core and high-volume genomic services are typically delivered through our local laboratory, a range of tests are provided through coordinated pathways within the NEY GLH. Some samples could also be referred to other GLH laboratories across the country, which may cover specialist services.
Below is an outline of our main testing modalities:
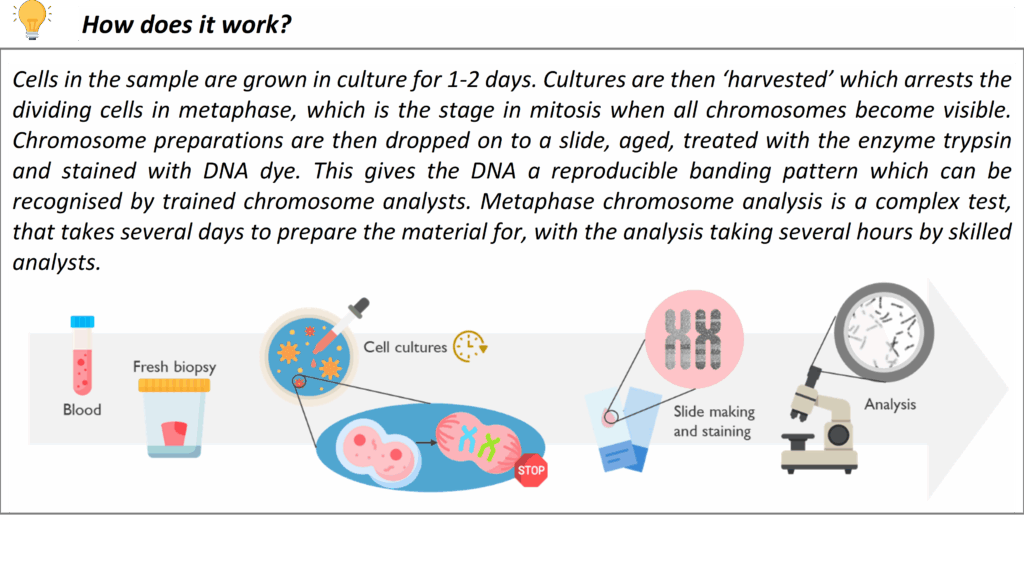
Karyotyping (Chromosome analysis)
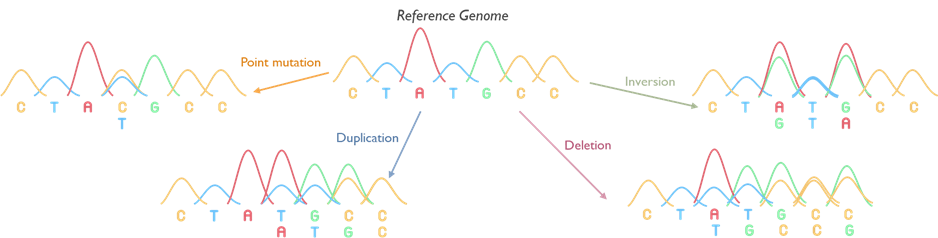
Karyotyping is the microscopic examination of chromosomes to detect large-scale genetic changes. This conventional G-banded chromosome analysis allows us to identify aneuploidies (extra or missing chromosomes) and structural rearrangements such as translocations, inversions, large deletions/duplications, and mosaicism.
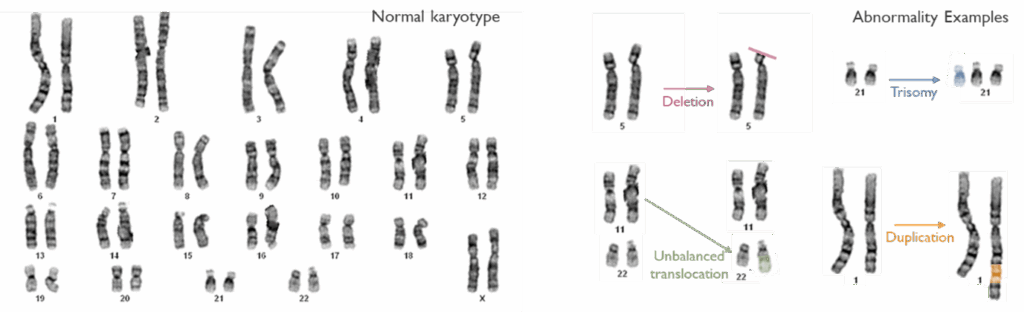
Karyotype analysis is offered for certain referral indications where a chromosomal abnormality is suspected – for example infertility, suspected balanced familial translocations and sex chromosome disorders. Karyotyping is also used as a reflex test to follow up on abnormalities detected by other methods: for instance, if a microarray shows an unbalanced chromosome segment, a karyotype of the patient or parents may be performed to characterize the rearrangement and assess inheritance risks. This test remains a valuable tool for understanding the whole genome at the chromosomal level, and for confirming findings like mosaic aneuploidy and balanced rearrangements that might be missed by other technologies.
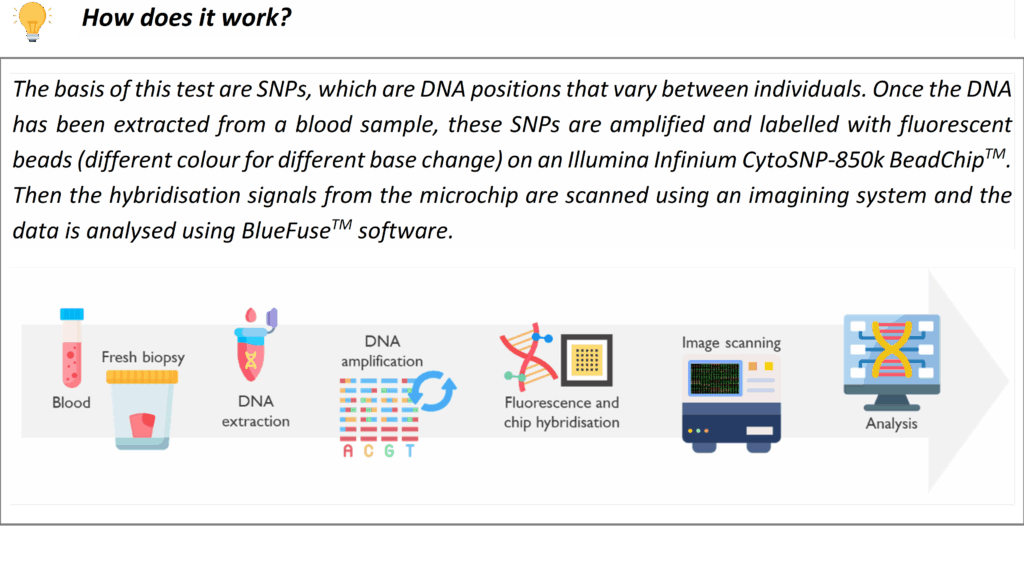
SNP (Single Nucleotide Polymorphism) arrays
SNP array is a high-resolution technique for detecting submicroscopic copy number variants (CNVs) across the genome. Our genome-wide SNP microarray can identify imbalance of DNA segments (deletions or duplications), that are too small to see on a karyotype. This is a first-line test for patients with global developmental delay, intellectual disability and/or congenital anomalies, as it can reveal pathogenic CNVs associated with syndromes. For example, microarray testing is commonly used to diagnose conditions like DiGeorge syndrome (22q11 deletion), Williams syndrome (7q11.23 deletion), or other microdeletion/microduplication syndromes. It is also utilized in prenatal diagnosis and pregnancy loss investigations: array testing can detect chromosome abnormalities that are causal of abnormal ultrasound findings or may explain the miscarriage or stillbirths (e.g. trisomy or unbalanced chromosomal rearrangements). Our laboratory follows national guidelines on when array testing is indicated for pregnancy losses. The SNP array also provides information on regions of homozygosity, which can be relevant for detecting uniparental disomy which is causal of some imprinting disorders.
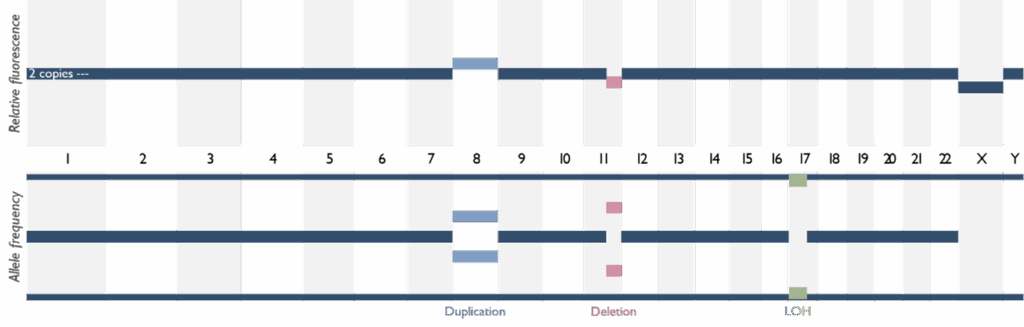
SNP arrays offer a significantly higher diagnostic yield than karyotype for many rare developmental disorders due to its ability to detect smaller imbalances. If the array identifies a genomic imbalance, our team will interpret its clinical significance in the context of the patient’s phenotype. Notably, if the array suggests the possibility of a single-gene disorder (for example, a deletion encompassing a specific gene), further targeted testing may be recommended.
Next-Generation Sequencing (NGS) Panels
We perform a broad menu of Next-Generation Sequencing panel tests, which allow simultaneous sequencing of multiple genes associated with rare disease phenotypes. These multi-gene panels are designed around clinical indications defined in the National Genomic Test Directory. For example, we have panels for Limb Girdle Muscular Dystrophy (LGMD), a group of neuromuscular disorders characterized by progressive muscle weakness and wasting, primarily affecting the muscles around the hips and shoulders. We also offer a breast cancer genetic testing panel that analyses multiple genes known to be associated with an increased risk of breast cancer, including BRCA1 and BRCA2, among others.
By sequencing all relevant genes in parallel, NGS panels can efficiently provide a genetic diagnosis for conditions that have genetic heterogeneity (multiple possible gene causes). Our panels use high-throughput sequencing technology to ensure deep coverage of target genes, and our bioinformatics pipeline filters variants to identify likely disease-causing mutations. Each gene panel test is validated and performed to clinical quality standards, and results are interpreted by our clinical scientists with input from clinicians as needed.
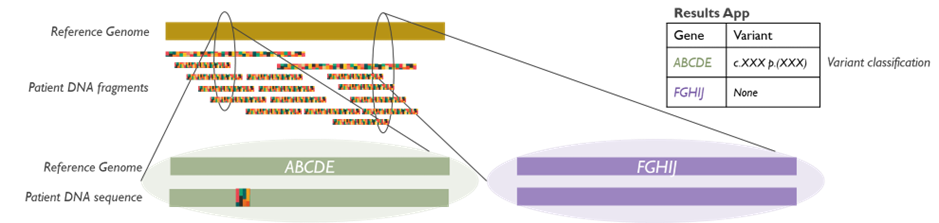
The introduction of NGS panels has greatly increased diagnostic yield for many rare diseases while reducing testing time and cost. For example, our inherited muscle disease panel can simultaneously check dozens of genes for muscular dystrophies (including LGMD subtypes) in a single test, rather than performing multiple individual gene tests sequentially. Similarly, our FH panel analyses the main genes known to cause familial hypercholesterolemia (such as LDLR, APOB, or PCSK9), allowing early identification of pathogenic variants so that patients and at-risk relatives can receive appropriate management. We continually update our panel content in line with the latest research and the Test Directory, and many panels have expanded to include newly discovered genes or have been retired in favour of whole genome analysis (see below).
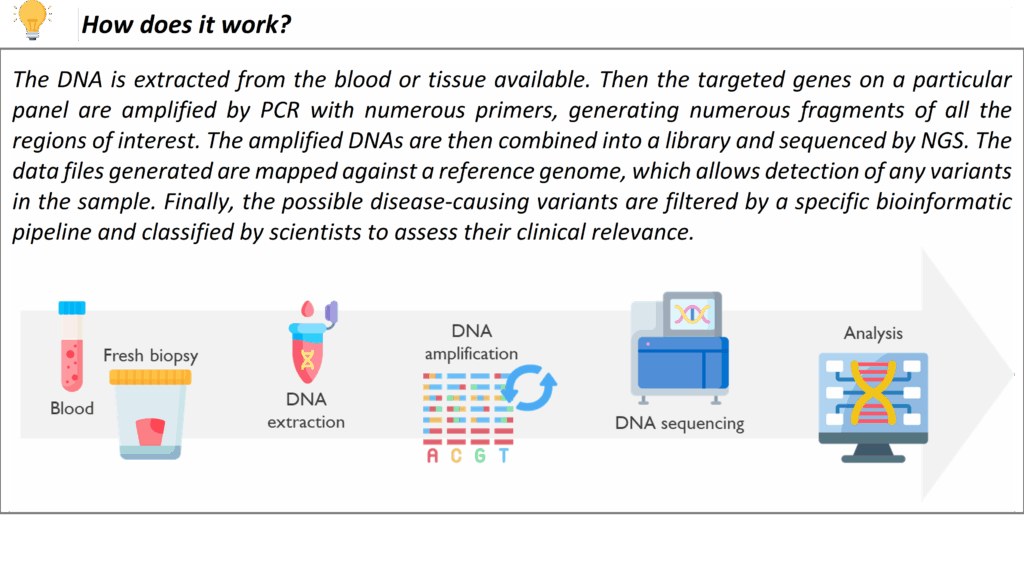
Whole Genome Sequencing (WGS)
Whole Genome Sequencing is the most comprehensive genomic test currently available, analysing the patient’s entire DNA sequence. Newcastle Genetics Laboratory is part of the national WGS programme commissioned by NHS England, and we offer WGS for patients who meet the eligibility criteria in the National Genomic Test Directory. Many rare disease indications have now moved to WGS in line with the evolution of the Genomic Medicine Service. For example, certain complex neurodevelopmental disorders, severe early-onset disorders, and cases where prior targeted testing (like gene panels) did not yield a diagnosis may now go straight to WGS. WGS can detect single nucleotide variants, small insertions/deletions, and copy number changes across the genome in one test, providing a higher chance of diagnosis for patients with elusive conditions.
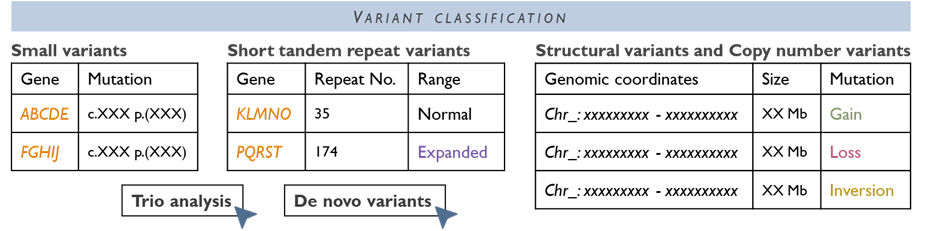
In our WGS pathway, samples are processed through a central pipeline with data generated at high coverage, and variant analysis is focused by phenotype-driven virtual gene panels (as per national protocol). We work closely with Genomics England and the NHS Central WGS pipeline to analyse and interpret the data. Where necessary, findings are reviewed in a multidisciplinary team (MDT) meeting to interpret the findings in the clinical context. Before WGS is initiated, patients (or their guardians) will have a discussion with their clinician and sign a Record of Discussion consent form, as WGS can generate additional information beyond the immediate clinical question. Our laboratory supports clinicians through this process. Once WGS is completed, a comprehensive report is issued. WGS has already helped diagnose previously unsolved cases, ending long diagnostic odysseys for some families, and it continues to be integrated for more conditions as the technology and evidence evolve.
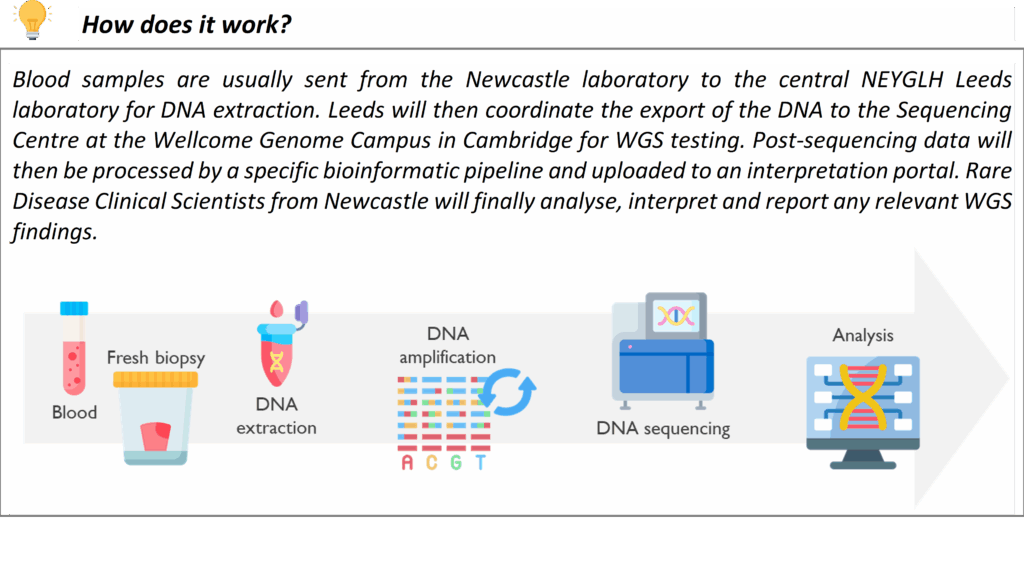
MassARRAY
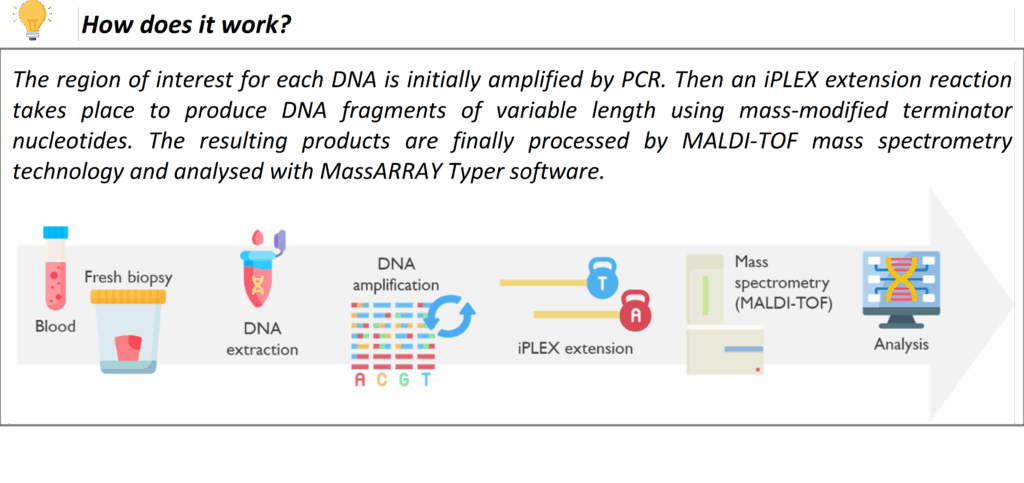
The MassARRAY is a high-throughput technique that uses MALDI-TOF Mass Spectrometry technology for rapid, targeted molecular testing. It enables the detection of DNA variants with high sensitivity. It is used for a variety of services within the Newcastle Genetics Laboratory, such as testing for the HFE gene mutations in Hereditary Haemochromatosis.
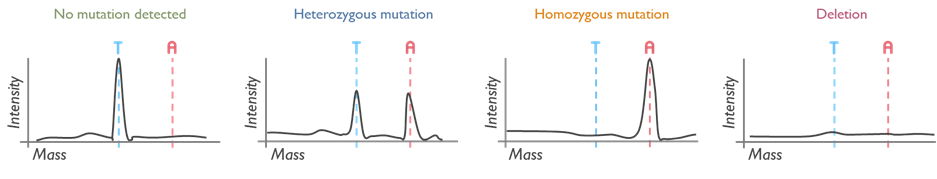
Its short turnaround time is extremely useful when used in newborn screening of rare and, when untreated, potentially severe disorders. MassARRAY detects the common ACADM variant that causes medium-chain acyl-CoA dehydrogenase deficiency (MCADD) and the common HBB variant in sickle cell disease in transfused infants. These tests are performed on DNA extracted from Guthrie spots (heel prick test), prenatal samples or blood.
A new service that our laboratory has taken recently from the Mitochondrial Laboratory is the detection of three variants commonly present in the MT-RNR1 gene, which confer susceptibility to deafness (ototoxicity) when exposed to aminoglycosides antibodies. This test is relevant for patients predisposed to infections or individuals with bilateral hearing loss who have been treated with aminoglycosides. Please contact the lab via [email protected] for any queries on this service.
Targeted Fragment Analysis
Targeted fragment length analysis (FLA) looks for a specific known variant or a small set of variants, rather than surveying an entire gene or genome, quantifying DNA fragments separated by size. This service is used for diagnostic testing (e.g. prenatal samples with high trisomy risk or abnormalities seen on scan), carrier testing (e.g. testing parents for a known familial cystic fibrosis mutation) and presymptomatic testing for adult-onset disorders (e.g. checking for a known familial mutation in the HTT gene for Huntington disease in an at-risk individual). Importantly, predictive or presymptomatic tests are only accepted if the individual has had proper counselling and meets criteria, often through Clinical Genetics referral.
We also perform repeat expansion analyses as targeted tests, for example, Huntington disease testing looks at the CAG repeat in the HTT gene. These specialized PCR-based tests determine the number of repeats to see if it falls in the affected range. Another example is testing for imprinting disorders via methylation-specific methods (e.g. Prader-Willi/Angelman syndrome methylation analysis at 15q11-q13).
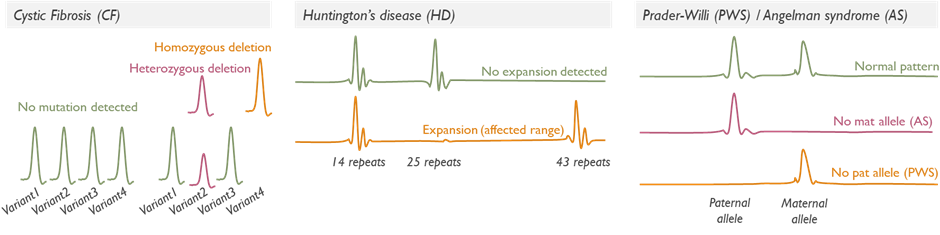
Our targeted tests are performed using the most appropriate technology for the variant in question. Turnaround for targeted tests is typically faster than for broad panels, given the smaller scope of analysis.
MLPA (Multiplex Ligation‑Dependent Probe Amplification)
MLPA is a technique we use to detect gene copy number changes (deletions or duplications of exons or whole genes) that are not easily found by sequencing or may be too small/regional for genome-wide arrays. MLPA involves using probe sets that hybridize to multiple specific targets in a gene or chromosomal region to determine if any targets are present in abnormal copy number.
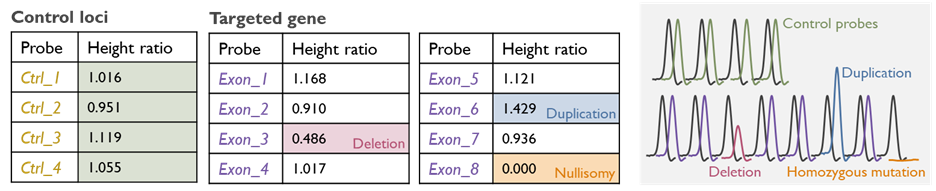
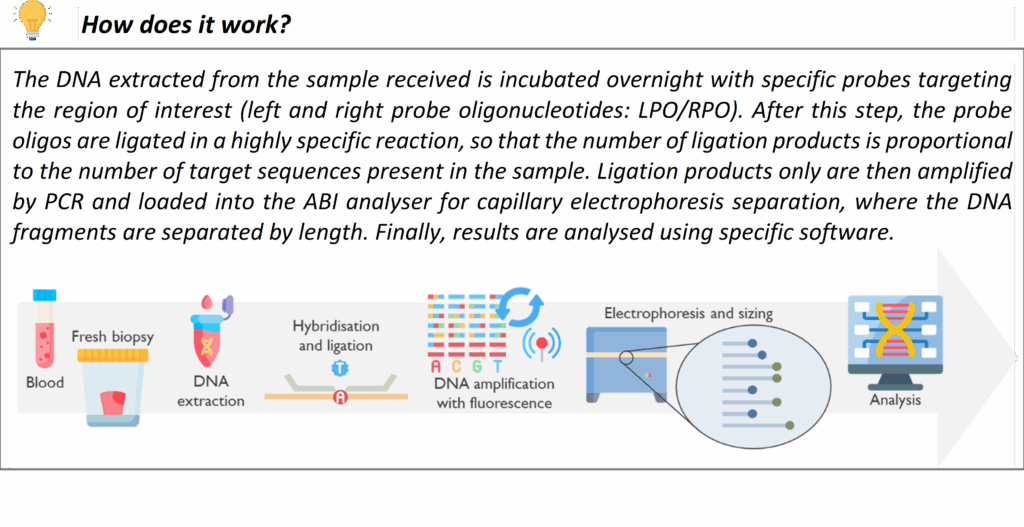
We utilize MLPA as a confirmatory and supplemental test for many conditions. For instance, certain genetic disorders commonly result from deletions or duplications of one or more exons in a gene e.g. PMP22 gene duplication/deletion in Charcot–Marie–Tooth disease type 1A/HNPP and spinal muscular atrophy (SMN1 gene deletion). Additionally, MLPA can be employed to confirm copy number findings from NGS or array data at a single-gene level when needed e.g. large deletions in the DMD gene causing Duchenne/Becker Muscular Dystrophy. This technology is an important part of our technology repertoire to ensure no pathogenic rearrangement is missed.
Droplet Digital PCR (ddPCR)
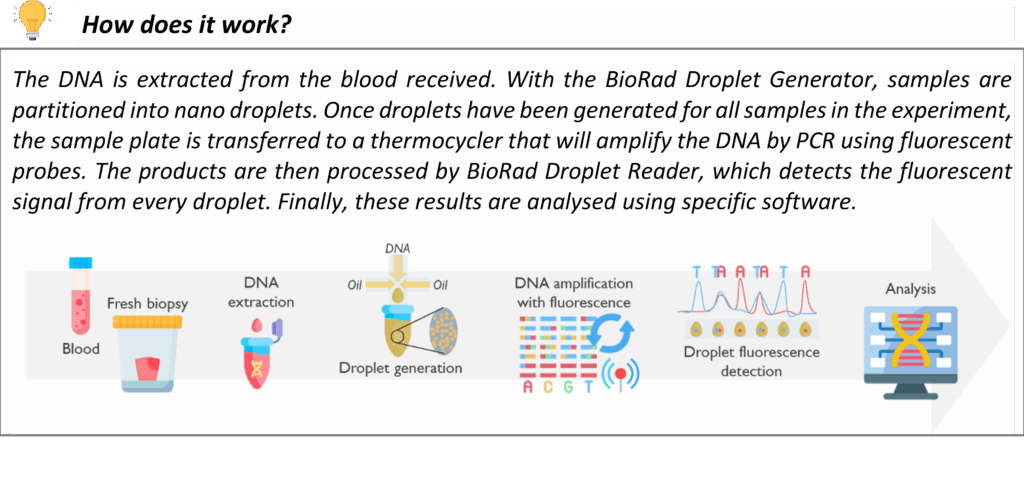
Droplet digital PCR is an advanced form of PCR that allows precise quantification of DNA variants or copy number by partitioning the sample into thousands of micro-reactions (droplets). In our rare disease service, we apply ddPCR in specialized situations that require a high level of sensitivity or quantitation.
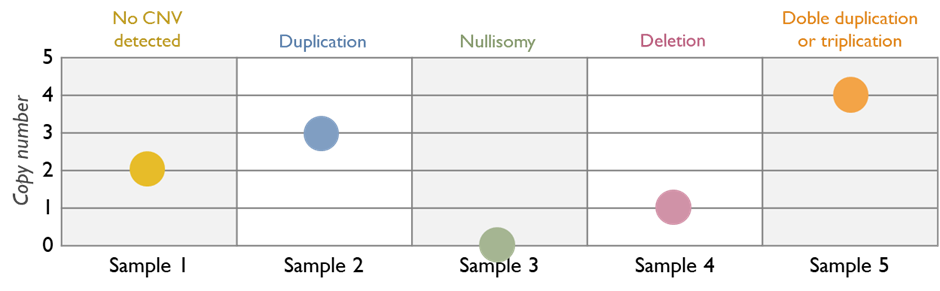
One application is for gene copy number confirmation similar to MLPA in certain scenarios, or for measuring transcript levels in research contexts. Whenever a quantitative DNA analysis is needed beyond the resolution of standard techniques, ddPCR provides an additional layer of confidence in the result. While not needed for most routine tests, this technology is available in our laboratory for those specific cases where it enhances diagnostic accuracy.
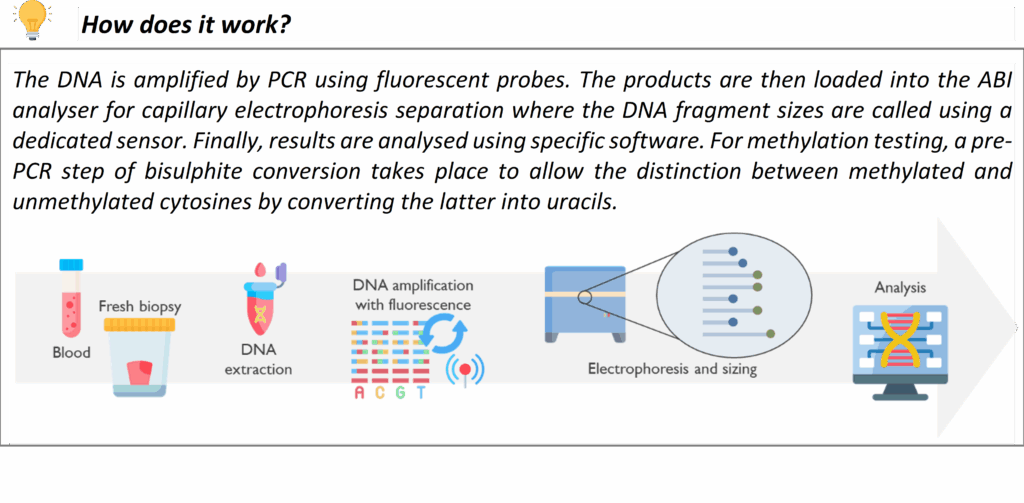
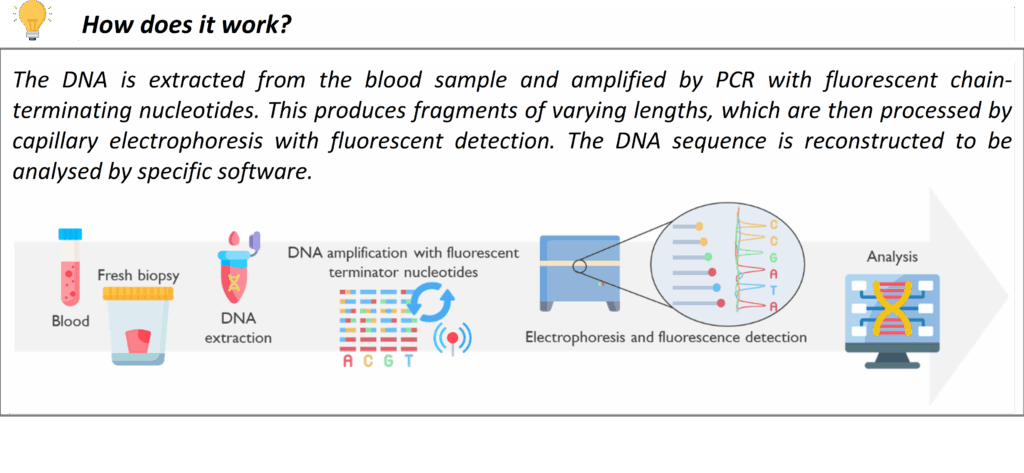
Sanger Sequencing (for Confirmation & Single-Gene Tests)
Sanger sequencing is a long-established DNA sequencing method that we continue to use as a complement to newer genomic techniques. In our laboratory, Sanger sequencing may be used to confirm key variants identified by NGS panel testing or WGS, or to resolve phase (determine if two mutations are on the same or different copies of a chromosome) in some cases by sequencing family members.
This service is crucial as well for cascade testing in families and for confirming known common pathogenic variants. If a pathogenic variant has been identified in a family (through one of the above methods or previously), we can offer predictive testing to relatives for that exact variant. For example, if a patient is found to have a disease-causing variant in the LDLR gene for Familial Hypercholesterolemia, we can perform targeted testing of that variant in siblings or children of the patient. This helps identify other family members who also have FH so they can receive early intervention. We offer similar cascade testing for many inherited conditions (in line with Clinical Genetics services). All Sanger sequencing in our lab is performed to diagnostic standards with appropriate controls.
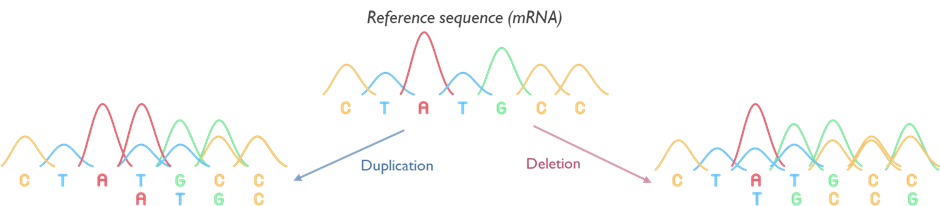
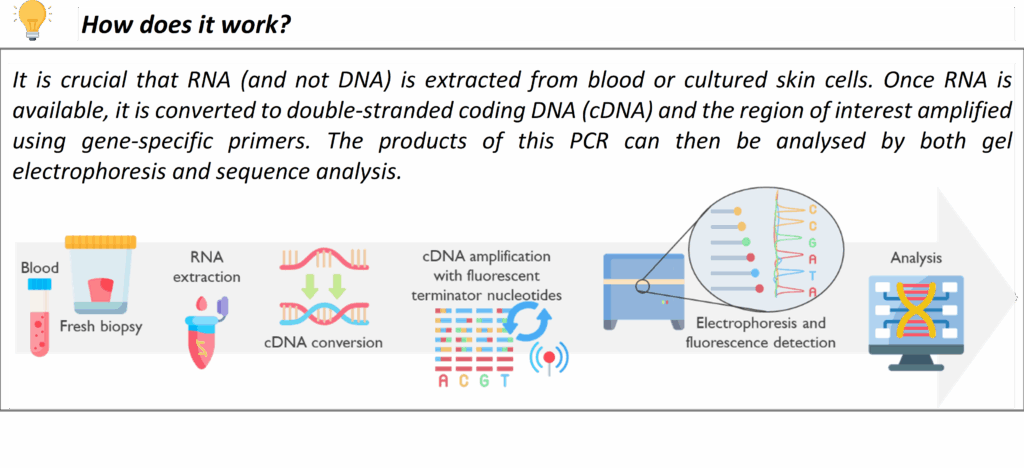
Targeted RNA analysis
Our laboratory offers a specialised targeted RNA sequencing service using the Sanger method, designed to deliver high-accuracy, single-transcript resolution. This approach is applied for the investigation of variants, identified through large scale (Panel / WGS screening), predicted to impact splicing and the results of these investigations are used to clarify the clinical significance of variants. This technique is used to evaluate splice transcripts at targeted sites to assess either the predicted disruption of putative splice sites or the introduction of novel splice site positions. Targeted Sanger RNA sequencing focuses on specific regions of interest, making it an ideal tool for cases where clear, base-level validation of transcript structure is essential.
In some cases, the impact of nonsense-mediated decay (NMD) can make the interpretation of these studies difficult. To overcome this, we can apply a temporary treatment that blocks NMD, allowing the RNA to stay intact that would otherwise be removed by that process. By including this step when needed, our service gives a more complete picture of what’s really going on inside the cell, supporting a more accurate assessment of any effect on splicing.
Please contact the lab via [email protected] if RNA studies is a line of investigation indicated for your patient.
How to Refer (Requesting Tests)
We welcome appropriate referrals from hospital specialists, clinical geneticists, and other appropriate healthcare professionals for patients with a suspected rare genetic condition. Users are directed to the Rare and inherited disease eligibility criteria detailing which patients should be considered for testing under that indication, and the requesting specialties who are eligible to order the test.
All requests for rare disease genomic tests must be submitted with the proper documentation to ensure we have the necessary information (please see our request forms). Key steps and requirements for referring to our service are outlined below:
- Test Request Forms: Use the standard Rare Disease genomic test request form for our laboratory. This form captures patient information, clinical details, and the specific test being requested. It is important to fill out the form completely, including the correct test R code (clinical indication code) and the test name/code as listed in the National Genomic Test Directory. Providing the Test Directory R code ensures that the request meets the national eligibility criteria and helps us direct the sample to the appropriate testing pathway. Please include detailed phenotypic information and family history where relevant, as this aids in interpretation. For certain specialised referrals, an additional proforma may be required, for example, there are specific templates for Familial Hypercholesterolemia, R14 Rapid Genome Sequencing Service, LGMD or aHUS referrals. Ensure these are attached if applicable.
- Whole Genome Sequencing referrals: If you are requesting WGS for a patient, please use the dedicated WGS test order form and indicate the WGS eligibility code from the National Genomic Test Directory. In addition, the national Record of Discussion (RoD) consent form for WGS must be completed with the patient/family prior to sending the sample. This form documents that the patient has been counselled about WGS and has given appropriate consent for the test, and for potential storage and future use of data. For trio WGS (testing a child and both parents), include details and samples for the parents as well. Guidance on completing the WGS paperwork or deciding if WGS is appropriate, can be found in the NEY GLH website.
- Sample requirements: For most rare disease genetic tests, the preferred sample is EDTA anti-coagulated blood (purple top tube). 3-5ml is sufficient in adults (or 1-2ml in infants), though larger volumes up to 10ml are welcome for extensive testing. We can accept genomic DNA (if extracted in another lab and of sufficient quality/quantity), prenatal samples (amniotic fluid or CVB), cord blood or tissue samples (e.g. skin biopsy for fibroblast culture, or products of conception in prenatal loss cases). Newborn screening cards (Guthrie cards) are not generally suitable for our tests due to DNA quantity. Buccal swabs may be also used for tests which require little DNA, like some targeted familial testing, although given their high failure rate, blood is preferred.
- For prenatal diagnosis of a known familial variant or specific condition, please contact us in advance to minimise the turnaround time. We typically require a foetal sample (chorionic villi or amniotic fluid) and parental samples for rapid testing to be arranged. Ensure all samples are clearly labelled with at least three patient identifiers (name, date of birth, NHS or hospital number) that match the prenatal request form. Improperly labelled samples or incomplete forms may result in rejection or delays (see our sample acceptance policy).
- Packaging and transport: Follow standard guidelines for packaging diagnostic specimens. Blood samples should be in a sealed biohazard bag with the form in a separate pocket. If mailing, pack with adequate cushioning and in compliance with UN3373 packaging instructions for biological samples. Avoid extreme temperatures – do not freeze blood samples. If samples cannot be shipped immediately, storing blood at 4°C for short durations (up to 24-48 hours) is acceptable.
- For RNA-based tests or other special requirements, please contact us for instructions [email protected]. Please use your local hospital courier system to send samples to Newcastle Genetics Laboratory, Central Parkway. If urgent, consider same-day courier or contact us to discuss logistics.
- Eligibility and clinical indication: By submitting a test request, the referrer is affirming that the patient meets the eligibility criteria for that genomic test as defined in the National Genomic Test Directory. Most rare disease tests have specific criteria (e.g. phenotype requirements, or that certain preliminary investigations have been done): please ensure these criteria are met and documented. If you are uncertain, refer to the Test Directory guidance or consult with a clinical geneticist. We do not offer testing for patients outside the NHS England criteria.
- Consent and data sharing: It is the responsibility of the referring clinician to obtain informed consent for genetic testing. Patients should be made aware that their sample will be used for the specific test and that remaining sample and/or DNA may be stored for future diagnostic need or quality assurance. As part of the Genomic Medicine Service, patients are also informed that anonymised data may be shared within the NHS and approved bodies for clinical audit, training, or research purposes, and that identifiable data might be shared if required by law (with appropriate safeguards). If a patient opts out of any aspect (for example, does not consent to DNA storage or to research use), please indicate this on the form or include the national opt-out form. Note that for WGS, the consent process covers these aspects in detail. We operate under the GDPR/Data Protection Act 2018 and maintain strict confidentiality of patient information.
- Referrals outside our laboratory scope: Newcastle Genetics Laboratory performs a wide array of tests, but if a required test is highly specialised and not offered here, we will facilitate referral of the sample to the appropriate accredited laboratory. For instance, certain Highly Specialised Services (such as complex metabolic genetics) might be provided by designated national laboratories. In such cases, send the sample and form to us as usual, and we will forward it with necessary paperwork to the specialist lab. You do not need to arrange this yourself. We coordinate within the Genomic Laboratory Hub network to ensure the test is done at the correct location. Results will be returned to you directly from the testing lab, and we will inform you of any such forwarding. This approach means you can refer to us for any genomic test in the Rare Disease National Genomic Test Directory, and we will either perform it or route it appropriately.
- Reporting of results: Once testing is complete, a report will be issued to the referring clinician (and any other copied recipients you specify, such as additional consultants or genetic counsellors). Reports include an interpretation of any clinically relevant variants identified, as well as recommendations for cascade testing or clinical management where appropriate. If no causative variant is found, the report will state that the result is negative or inconclusive, and may still include relevant information (for example, carrier status for other recessive conditions may be reported if detected, according to consent and policy). We endeavour to make our reports clear and informative for non-geneticists. If you require assistance interpreting a report or discussing next steps, our clinical scientists and the Clinical Genetics team are available for discussion.
Turnaround Times
We understand that timely results are crucial for patient care. Our laboratory works to meet or shorten the turnaround times (TAT) targets set by NHS England for genomic tests. Actual turnaround times vary depending on the complexity and urgency of the test. Please refer to our Genetics Turnaround Times document to check the aimed reporting timeframes for rare disease services.
Please note that turnaround times count from the date the sample and completed paperwork are received in our laboratory. Incomplete information or the need for additional samples can extend the process, so providing thorough information upfront helps us keep within expected timeframes. We encourage clinicians to reach out if a result is needed urgently for clinical reasons, we will do our best to accommodate and prioritise that case.
We provide the latest service information via our website updates section. If there are any unforeseen delays (e.g. equipment maintenance, need for a sample recollection, or external referral delays), which might affect patient management, we will contact the referring clinician directly.
Contact Us
If you have any questions about our rare disease genomic services, need advice on test selection and interpretation, or issues related to sample logistics (e.g. if you need to arrange a special courier or have a question about sample stability) please refer to the FAQ section the first instance. If you query remains outstanding, please do not hesitate to contact the Newcastle Genetics Laboratory:
Newcastle Genetics Laboratory
Biomedicine East, Centre for Life
Central Parkway
Newcastle upon Tyne
NE1 3BZ
Telephone (Enquiries): 0191 241 8775 (service manager)
Email: [email protected] (General laboratory email for rare disease genomics enquiries)
We are available Monday to Friday, 9:00 am – 5:00 pm for telephone enquiries. Routine queries can also be sent via email; our team monitors the inbox during business hours and will respond as promptly as possible.
Additional Contacts: If your query is about a clinical genetics appointment or referral (as opposed to laboratory testing), please contact the Clinical Genetics Service separately at [email protected].
Multidisciplinary teams (MDT)
For clinicians within our network, we also offer the option of discussing cases at our Genomic multidisciplinary team (MDT) meetings, please contact us if you wish to present a complex case or review results in a multidisciplinary forum.
We value feedback on our services. If you have any suggestions, concerns, or compliments, feel free to reach out. There is a feedback form on our website, or you can email the laboratory manager directly. Our goal is to provide a world-class genomic service for rare diseases, and we continuously seek to improve in partnership with our users.
FAQs
Sample requirements
What is the appropriate sample container for genetic testing?